| Site1 | |
|---|---|
| Sp. 1 | 4 |
| Sp. 2 | 300 |
| Sp. 3 | 56 |
| Sp. 4 | 23 |
Diversity and Trait-Based Approaches
🧑🏻💻 Masatoshi Katabuchi @ XTBG, CAS
- mattocci27@gmail.com
- mattocci.bsky.social
- github.com/mattocci27/phy-fun-div
- https://mattocci27.github.io
November 18-20, 2025 XTBG AFEC
Objective
We Learn:
Why we use functional and phylogenetic diversity
How to calculate functional and phylogenetic diversity
Trait-based approaches
Outline
Community Ecology
Simple diversity indices
Phylogenetic diversity
Functional traits and diversity
R examples
Community Assembly and Species Coexistence
For over a century, field ecologists have been characterizing patterns in ecological communities and trying to draw theoretical inferences from the resulting data.

Central Questions:
Why do species occur in specific locations?
Why do some species coexist while others do not?
- Environmental filtering:
- Ecologically similar species should coexist in ecologically similar environments.
- Limiting similarity:
- Ecologically dissimilar species should coexist because too similar species competing for the same resources cannot stably coexist.
- Neutral theory:
- Dispersal and stochastic demographic processes explain species coexistence and species differences are not important.
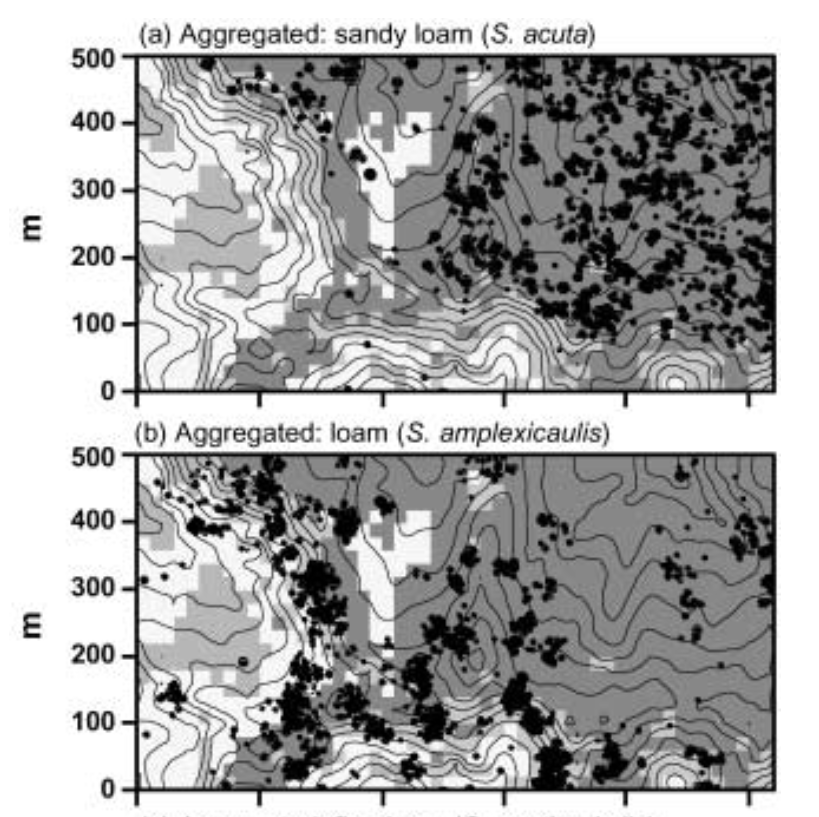
Russo et al. 2005, Journal of Ecology.
How can we quantify ecological similarity of coexisting species?
How to quantify ecological communities 🌿
Species
Species + Site information (1950s ~)
Species + Site information + Species information (2000s ~)
1a) First-order properties of single communities
[1] Vellend, M. (2016). The Theory of Ecological Communities. Princeton University Press
A vector of species abundance
Species composition
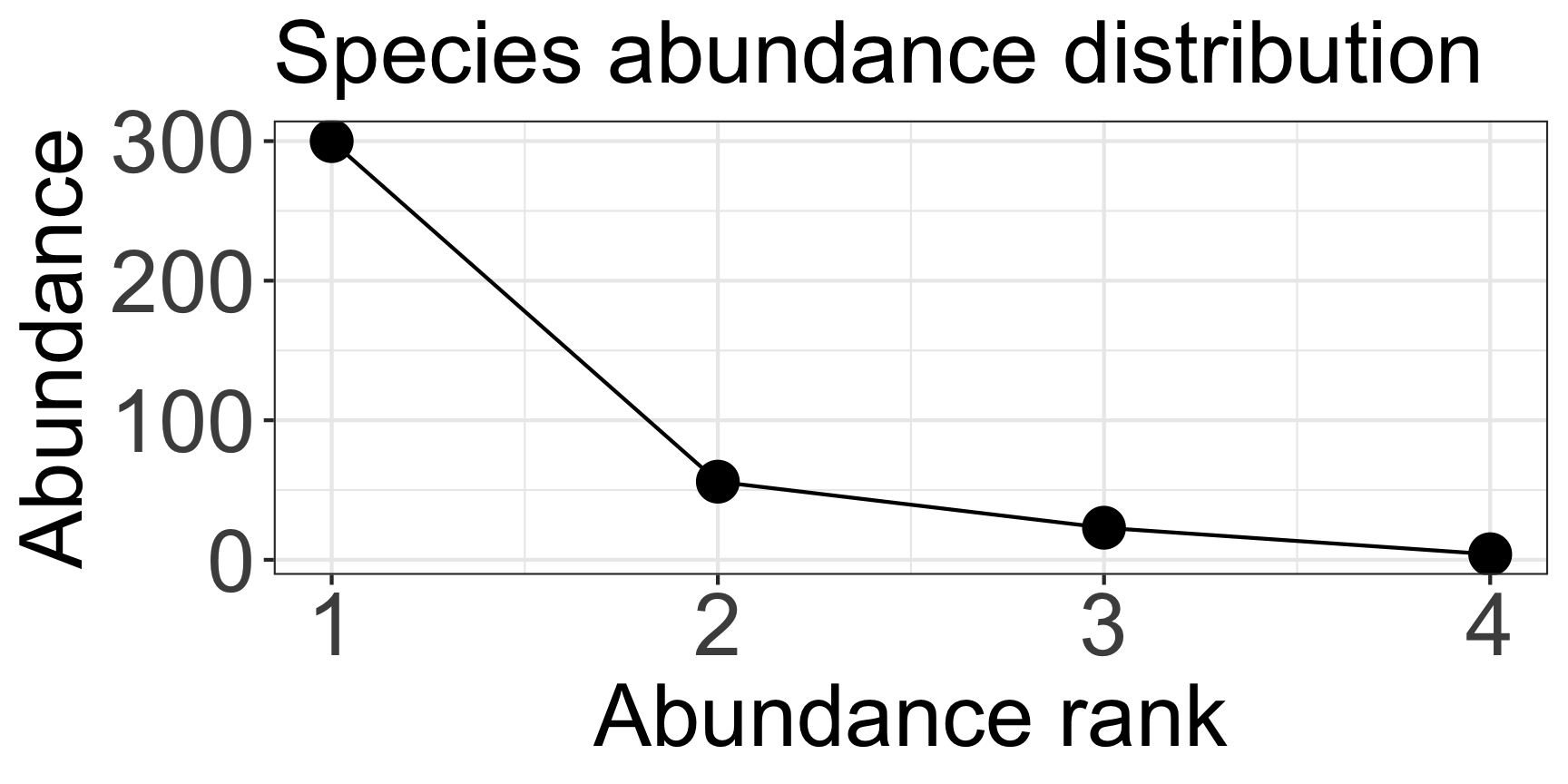
- Species richness = 4
- Simpson’s evenness = 1/ Σfreqi2 = (4/383)2 + (300/383)2 + (56/383)2 + (23/383)2
1a) First-order properties of single communities
[1] Vellend, M. (2016). The Theory of Ecological Communities. Princeton University Press
Which community is more diverse?
Species richness = 2
What is the chance to get the same species?
A: \(\frac{9}{10} \times \frac{8}{9} + \frac{1}{10} \times \frac{0}{9} = 0.8\)
B: \(\frac{5}{10} \times \frac{4}{9} + \frac{5}{10} \times \frac{4}{9} \simeq 0.44\)
1a) First-order properties of single communities
Which community is more diverse?

A: \(\frac{9}{10} \times \frac{8}{9} + \frac{1}{10} \times \frac{0}{9} = 0.8\)
B: \(\frac{5}{10} \times \frac{4}{9} + \frac{5}{10} \times \frac{4}{9} \simeq 0.44\)
We prefer that large values indicate more diverse communities.
Diversity of A: 1 - 0.8 = 0.2
Diversity of B: 1 - 0.44 = 0.56
Simpson’s Index of Diversity: \(D = 1 - \Sigma\frac{n_i(n_i - 1)}{N_i(N_i - 1)}\)
Simpson’s Index of Diversity (ver. 2): \(D = 1 - \Sigma p_i^2\)
n: number of individuals of each species, N: total number of individuals of all species, p: relative species abundance
[1] Vellend, M. (2016). The Theory of Ecological Communities. Princeton University Press
1a) First-order properties of single communities
Another simple way to describe diversity?

A: \(p_1\) = 0.9, \(p_2\) = 0.1
B: \(p_1\) = 0.5, \(p_2\) = 0.5
Diversity of A: 0.9 \(\times\) 0.1 = 0.09?
Diversity of B: 0.5 \(\times\) 0.5 = 0.25?
Diversity \(\times\) Diversity? What is the unit?
- \(\mathrm{log}(x \times y) = \mathrm{log}(x) + \mathrm{log}(y)\)
- Expectations:
- A: \(0.9 \times \mathrm{log}(0.9) + 0.1 \times \mathrm{log}(0.1) \simeq -0.32\)
- B: \(0.5 \times \mathrm{log}(0.5) + 0.5 \times \mathrm{log}(0.5) \simeq -0.69\)
- We prefer that large values indicate more diverse communities.
- A: \(-1 \times (-0.32) = 0.32\)
- B: \(-1 \times (-0.69) = 0.69\)
- Shannon Diversity Index: \(H' = -\Sigma p_i\mathrm{log}p_i\)
p: relative species abundance
[1] Vellend, M. (2016). The Theory of Ecological Communities. Princeton University Press
1b) First-order properties of multiple communities (Beta diversity)
[1] Vellend, M. (2016). The Theory of Ecological Communities. Princeton University Press
- Species \(\times\) site matrix
- Metacommunity
| Site 1 | Site 2 | Site 3 | Site 4 | |
|---|---|---|---|---|
| Sp. 1 | 4 | 0 | 315 | 23 |
| Sp. 2 | 300 | 250 | 0 | 18 |
| Sp. 3 | 56 | 120 | 74 | 0 |
| Sp. 4 | 23 | 18 | 101 | 0 |
- Dissimilarity matrix (site \(\times\) site)
| Site 1 | Site 2 | Site 3 | Site 4 | |
|---|---|---|---|---|
| Site 1 | 0.00 | 0.16 | 0.81 | 0.90 |
| Site 2 | 0.16 | 0.00 | 0.79 | 0.92 |
| Site 3 | 0.81 | 0.79 | 0.00 | 0.91 |
| Site 4 | 0.90 | 0.92 | 0.91 | 0.00 |
e.g., Bray–Curtis dissimilarity
\(BC_{ij}=1-2\frac{\sum min\left(S_{A,i}\mbox{, } S_{B,i}\right)}{\sum S_{A,i}+\sum S_{B,i}}\)
Site 1 vs Site 2: 1 - (2 * (0 + 250 + 56 + 18) / (4 +300 + 56 + 23 + 0 + 250 + 120 + 18)) = 0.16
2a) Second-Order properties with site characteristics (1950s ~)
[1] Vellend, M. (2016). The Theory of Ecological Communities. Princeton University Press
| Site 1 | Site 2 | Site 3 | Site 4 | |
|---|---|---|---|---|
| Abundance | ||||
| Sp. 1 | 4 | 0 | 315 | 23 |
| Sp. 2 | 300 | 250 | 0 | 18 |
| Sp. 3 | 56 | 120 | 74 | 0 |
| Sp. 4 | 23 | 18 | 101 | 0 |
| Env | ||||
| Env. 1 | 780 | 2500 | 480 | 1200 |
| Env. 2 | 21 | 11 | 24 | 19 |
| Env. 3 | 1500 | 1900 | 700 | 4500 |
- “Species \(\times\) site” and “site \(\times\) environment”
2a) Second-Order properties with site characteristics (1950s ~)
[1] Vellend, M. (2016). The Theory of Ecological Communities. Princeton University Press
| Site 1 | Site 2 | Site 3 | Site 4 | |
|---|---|---|---|---|
| Abundance | ||||
| Sp. 1 | 4 | 0 | 315 | 23 |
| Sp. 2 | 300 | 250 | 0 | 18 |
| Sp. 3 | 56 | 120 | 74 | 0 |
| Sp. 4 | 23 | 18 | 101 | 0 |
| Env | ||||
| Elevation (m) | 780 | 2500 | 480 | 1200 |
| MAT (℃) | 21 | 11 | 24 | 19 |
| MAP (mm) | 1500 | 1900 | 700 | 4500 |
- “Species \(\times\) site” and “site \(\times\) environment”
- Diversity-environment relationships
- Composition-environment relationships
- Multivariate ordination: placing the survey plots “in order” based on their multivariate species composition.
2b) Second-Order properties with species characteristics (2000s ~)
[1] Vellend, M. (2016). The Theory of Ecological Communities. Princeton University Press
| Site 1 | Site 2 | Site 3 | Site 4 | |
|---|---|---|---|---|
| Abundance | ||||
| Sp. 1 | 4 | 0 | 315 | 23 |
| Sp. 2 | 300 | 250 | 0 | 18 |
| Sp. 3 | 56 | 120 | 74 | 0 |
| Sp. 4 | 23 | 18 | 101 | 0 |
| Env | ||||
| Elevation (m) | 780 | 2500 | 480 | 1200 |
| MAT (℃) | 21 | 11 | 24 | 19 |
| MAP (mm) | 1500 | 1900 | 700 | 4500 |
| Trait 1 | Trait 2 | Trait 3 | Trait 4 | |
|---|---|---|---|---|
| Sp 1 | 1.3 | 4.8 | 1.8 | 30.0 |
| Sp 2 | 1.7 | 12.5 | 2.1 | 22.4 |
| Sp 3 | 7.0 | 5.9 | 5.7 | 11.5 |
| Sp 4 | 2.1 | 2.1 | 3.4 | 119.9 |
- “Trait \(\times\) species”, “species \(\times\) site”, “site \(\times\) environment”
2b) Second-Order properties with species characteristics (2000s ~)
[1] Vellend, M. (2016). The Theory of Ecological Communities. Princeton University Press
| Site 1 | Site 2 | Site 3 | Site 4 | |
|---|---|---|---|---|
| Abundance | ||||
| Sp. 1 | 4 | 0 | 315 | 23 |
| Sp. 2 | 300 | 250 | 0 | 18 |
| Sp. 3 | 56 | 120 | 74 | 0 |
| Sp. 4 | 23 | 18 | 101 | 0 |
| Env | ||||
| Elevation (m) | 780 | 2500 | 480 | 1200 |
| MAT (℃) | 21 | 11 | 24 | 19 |
| MAP (mm) | 1500 | 1900 | 700 | 4500 |
| Leaf N | Amax | Rdark | LL | |
|---|---|---|---|---|
| Sp 1 | 1.3 | 4.8 | 1.8 | 30.0 |
| Sp 2 | 1.7 | 12.5 | 2.1 | 22.4 |
| Sp 3 | 7.0 | 5.9 | 5.7 | 11.5 |
| Sp 4 | 2.1 | 2.1 | 3.4 | 119.9 |
- “Trait \(\times\) species”, “species \(\times\) site”, “site \(\times\) environment”
Trait diversity and its role in species composition
Trait composition-environment relationships
How to measure species characteristics?
Photosynthetic rates

Assuming closely related species are more ecologically similar
- Genus:species ratio: Relatedness as a substitute for ecological similarity
Community with 1 genus and 3 species (1:3)
Community with 3 genus and 3 species (3:3)
A low genus:species ratio indicates closely related species coexist.
- Environmental filtering
A high genus:species ratio indicates distantly related species coexist.
- Limiting similarity
Assuming closely related species are more ecologically similar
- Genus:species ratio: Relatedness as a substitute for ecological similarity
Community with 3 genus and 3 species (3:3)

Community with 3 genus and 3 species (3:3)

A low genus:species ratio indicates closely related species coexist.
- Environmental filtering?
A high genus:species ratio indicates distantly related species coexist.
- Limiting similarity?
Genus:species ratio
[1] Swenson 2013, Ecography
- The genus:species ratio type of study in plant community ecology started ~1910 and was popular until 1990’s
- A large criticism of genus:species ratio analyses is that they do not take account for the different ages of genera and species
- Two species in a relatively young genus may be expected to be more similar than two species in a relatively old genus.
Solution for the genus:species ratio problem = Use phylogenetic trees
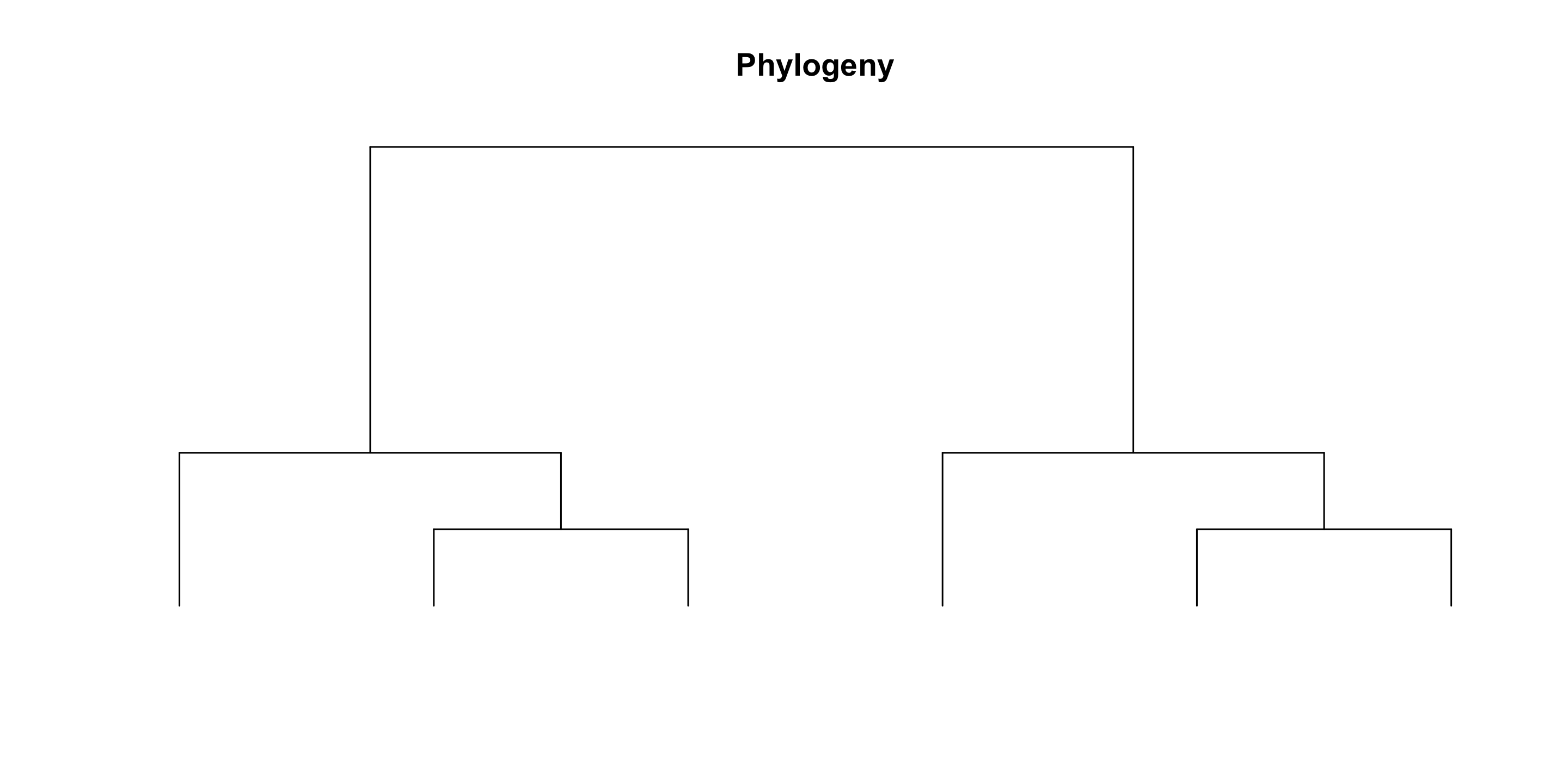
Phylodiversity
[1] Dimensions of Biodiversity, NSF
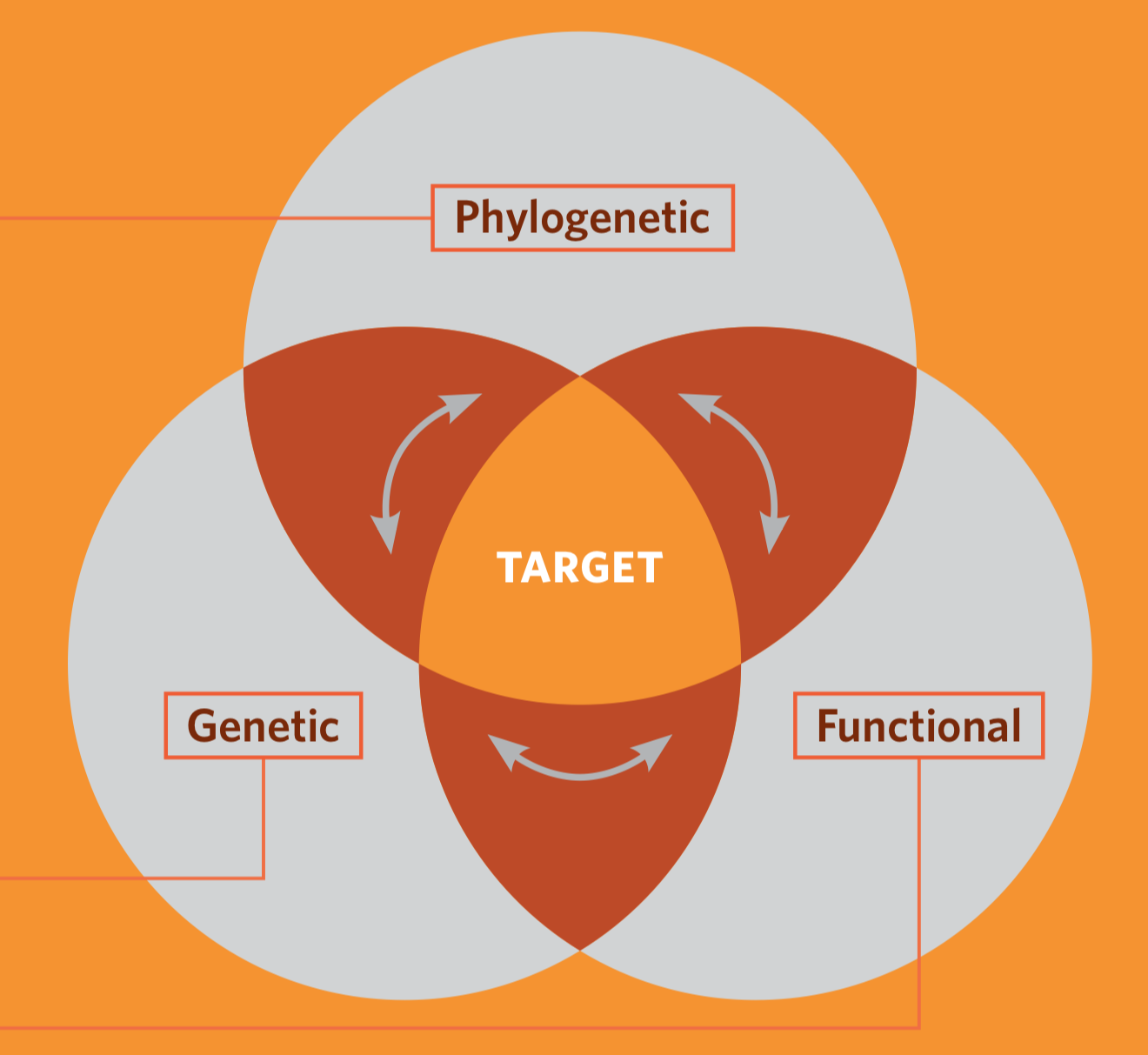
In the 1990’s conservation biologists recognized the biodiversity is not only species diversity
- Biodiversity has several axes or dimensions including genetic, taxonomic, phylogenetic and functional diversity
Phylodiversity
[1] Faith 1992, Biological Conservation
- Phylogenetic diversity was first formalized by Dan Faith in 1992
- He proposed a metric called PD that is also commonly referred to as Faith’s Index
- Many additional metrics have now been generated but this metric is still widely used, especially in the context of conservation
Faith’s Index (PD)
[1] Faith 1992, Biological Conservation
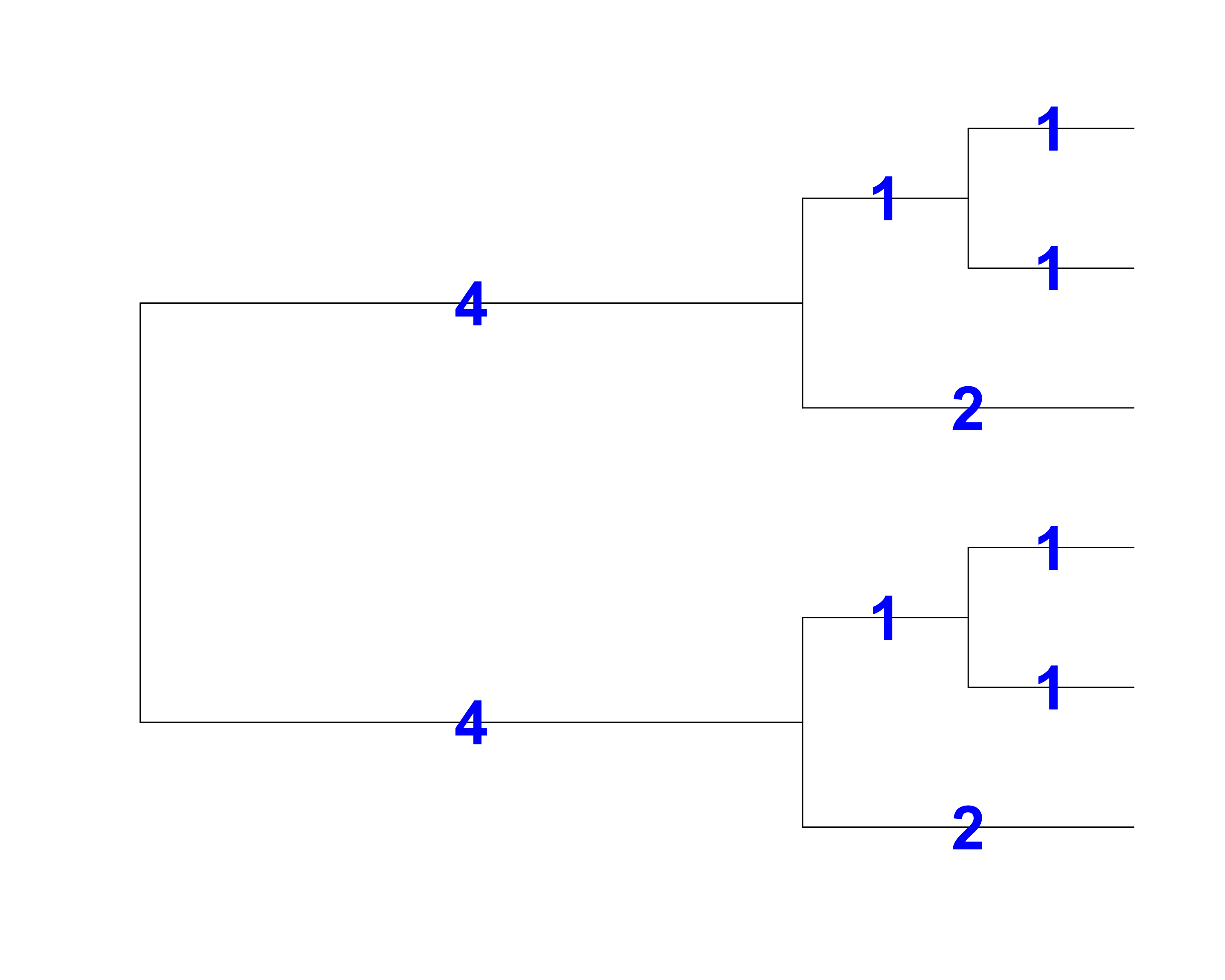
- Total branch length = 18
- PD is the sum of the lengths of all those branches that are members of the corresponding minimum spanning path
- PD is the phylogenetic analogue of taxon richness and is expressed as the number of tree units which are found in a sample
- PD will correlate with species richness
Faith’s Index (PD)
[1] Faith 1992, Biological Conservation
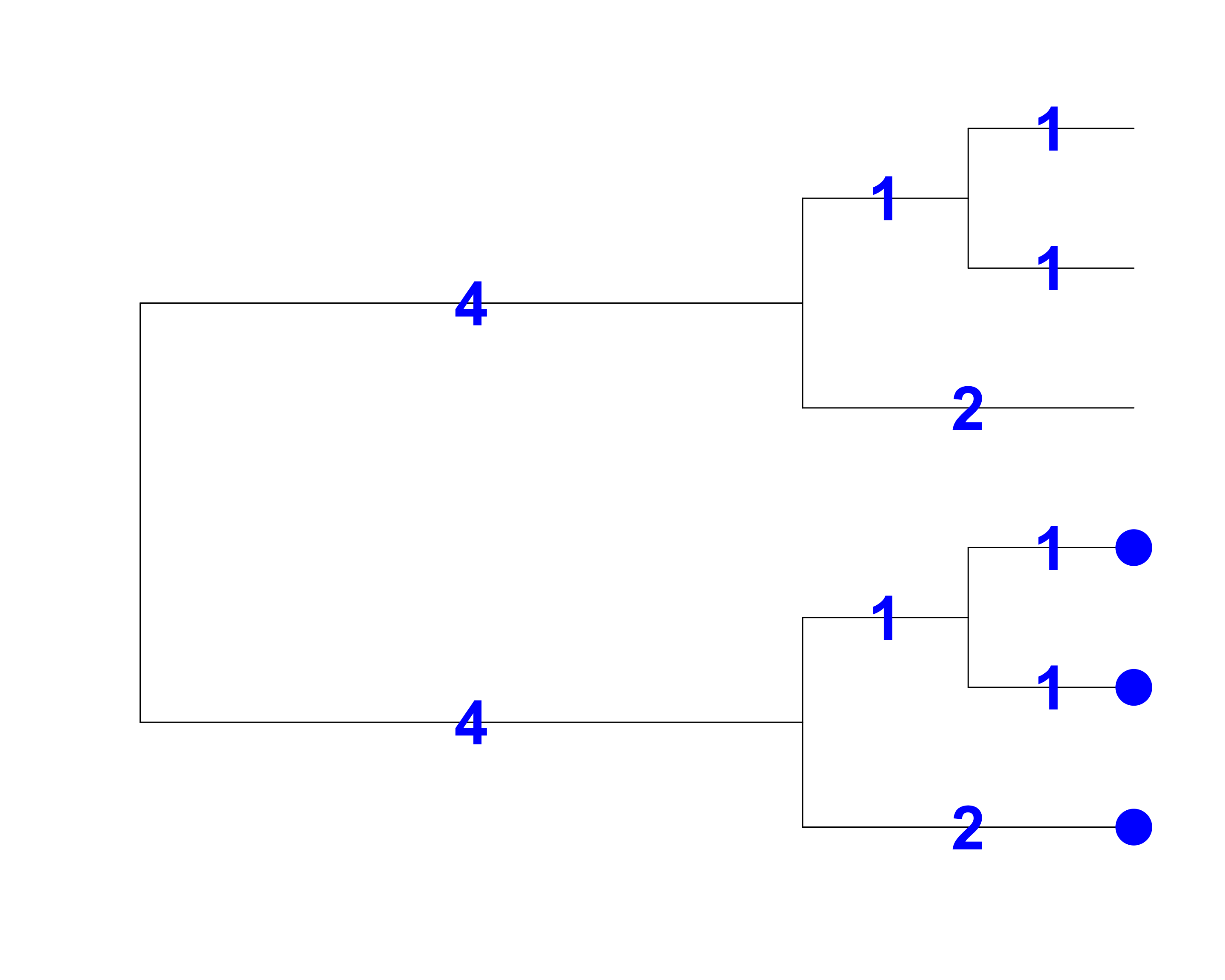
- Total branch length = 9
Faith’s Index (PD)
[1] Faith 1992, Biological Conservation
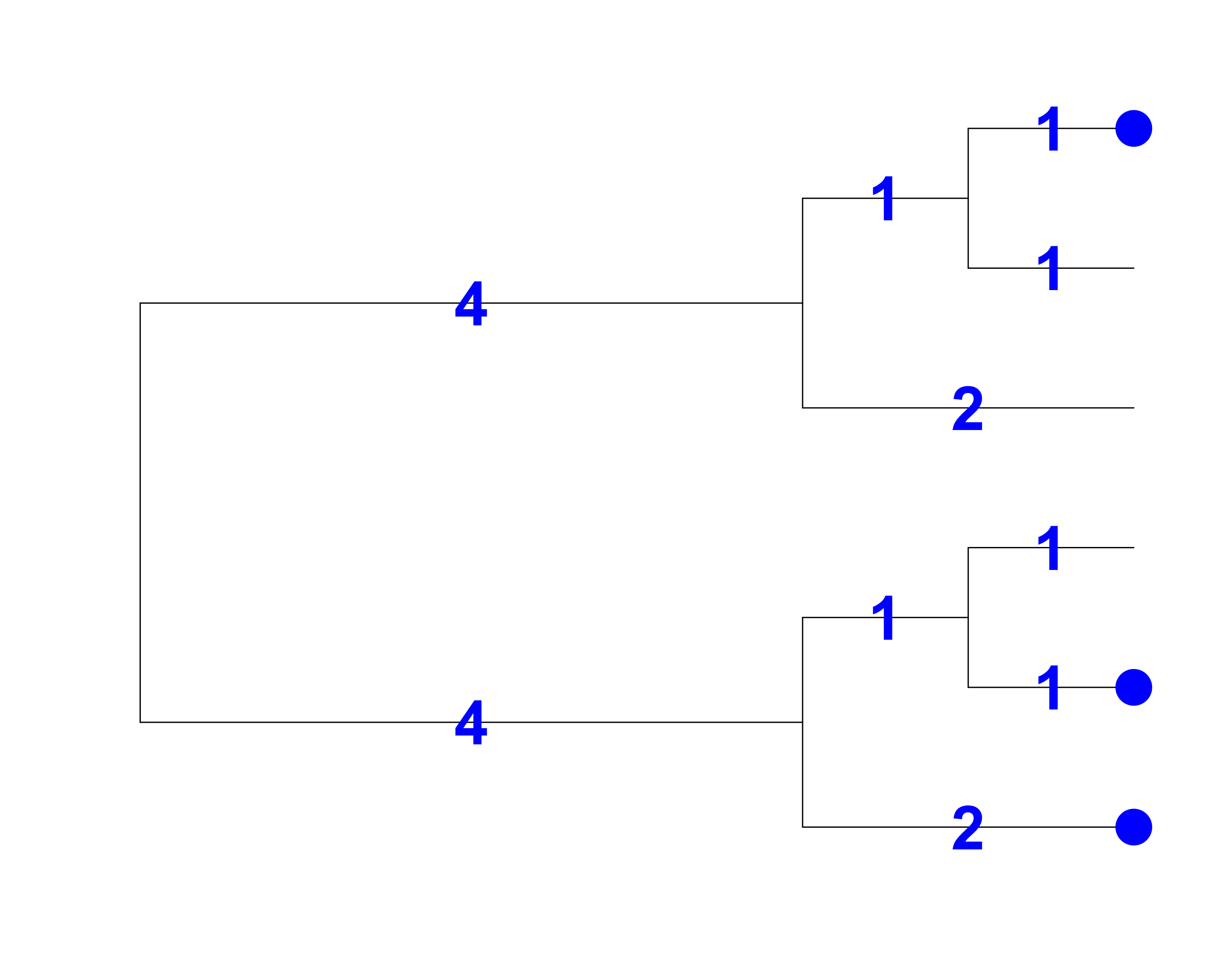
- Total branch length = 14
Petchey’s functional diversity (FD)
[1] Petchey & Gaston 2002, Ecology Letters
- FD is proposed by Owen Petchey in 2002
- FD is the total branch length of the functional dendrogram.
- Analogous to PD
Beyond Faith’s Index (PD)
[1] Webb 2000, The American Naturalist
Solution for genus:species = Use phylogenetic trees to estimate the relatedness of coexisting species
- This solution was first proposed by Cam Webb in 2000
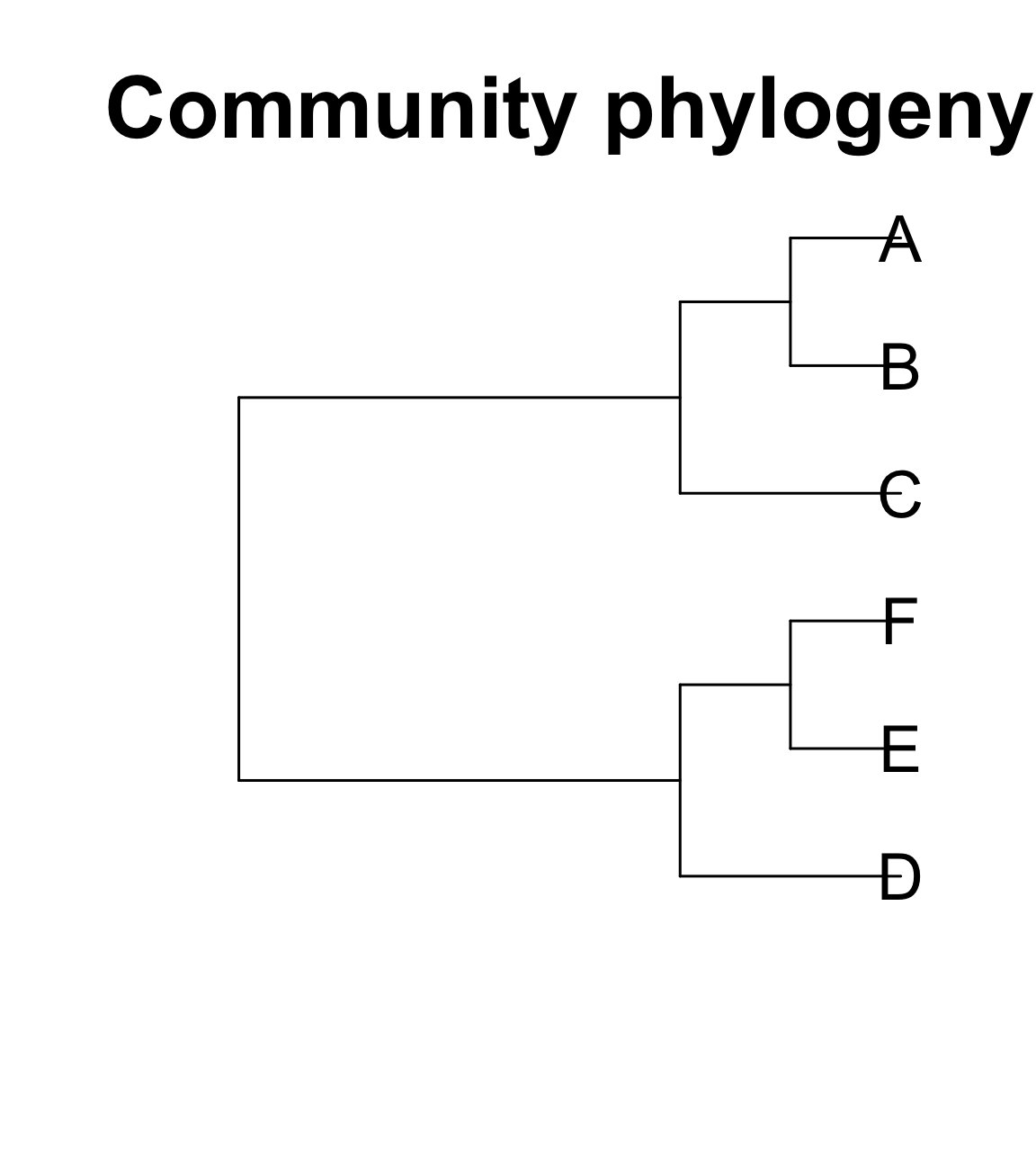
Distance matrix
A B C D E
B 1
C 2 2
D 4 4 3
E 5 5 4 2
F 5 5 4 2 1Mean Pairwise Distance (MPD) and Net Related Index (NRI)
[1] \(MPD = \frac{1}{n} \sum^{n}_{i} \sum^n_j \delta_{i,j} \; i \neq j\), where \(\delta_{i, j}\) is the pairwise distance between species i and j
Let’s consider greatest possible mean pairwise node distance (MPD) for a community of 4 taxa

A B C D E
B 1
C 2 2
D 4 4 3
E 5 5 4 2
F 5 5 4 2 1 A B E
B 1
E 5 5
F 5 5 1Greatest MPD for a community of 4 taxa: 22 / 6 pairs = 3.66 (A, B, E, F)
Mean Pairwise Distance (MPD) and Net Related Index (NRI)
[1] \(MPD = \frac{1}{n} \sum^{n}_{i} \sum^n_j \delta_{i,j} \; i \neq j\), where \(\delta_{i, j}\) is the pairwise distance between species i and j
Greatest possible mean pairwise node distance for a community of 4 taxa: 22 / 6 pairs = 3.66 (A, B, E, F)
Community 1; A, B, C, D
A B C
B 1
C 2 2
D 4 4 3MPD = (1 + 2 + 2 + 4 + 4 + 3) / 6 = 2.66
NRI = 1 - (2.66 / 3.66) = 0.273
Community 2; A, B, E, F
A B E
B 1
E 5 5
F 5 5 1MPD = (1 + 5 + 5 + 5+ 5 + 1) / 6 = 3.66
NRI = 1 - (3.66 / 3.66) = 0
Community 1 is more phylogenetically similar.
Mean Nearest Nodal Distance (MNTD) and Nearest Taxa Index (NTI)
[1] \(MNTD = \frac{1}{n} \sum^n_i min \delta_{i,j} \; i \neq j\), where \(min \delta_{i, j}\) is the minimum distance between species i and all other species in the community.
Greatest possible nearest nodal distance for a community of 4 taxa = 2 (A, C, D, F; A to C = 2, D to F = 2)

A B C D E
B 1
C 2 2
D 4 4 3
E 5 5 4 2
F 5 5 4 2 1Mean Nearest Nodal Distance (MNTD) and Nearest Taxa Index (NTI)
[1] \(MNTD = \frac{1}{n} \sum^n_i min \delta_{i,j} \; i \neq j\), where \(min \delta_{i, j}\) is the minimum distance between species i and all other species in the community.
Community 1; A, B, C, D
- A -> B
- B -> A
- C -> (A, B)
- D -> C
A B C
B 1
C 2 2
D 4 4 3MNTD = (1 + 1 + 2 + 3) / 4 = 1.75
NTI = 1 - (1.75 / 2.0) = 0.125
Community 2; A, B, E, F
- A -> B
- B -> A
- E -> F
- F -> E
A B E
B 1
E 5 5
F 5 5 1MNTD = (1 + 1 + 1 + 1) / 4 = 1
NTI = 1 - (1 / 2.0) = 0.5
Community 2 is more phylogenetically similar (in tips).
Sparks community phylogeny

Do phylogenetically related species have similar ecological niches?
[1] Anolis ecomorphs (Wikipedia)

We are assuming that related species are ecologically similar
Related species sometimes have very different traits and ecological niches (e.g., grass-bush, trunk, trunk-crown, trunk-ground and twig ecomorphs)
Functional dendrogram vs. phylogeny (Anole example)
[1] Losos et al. 1998, Science
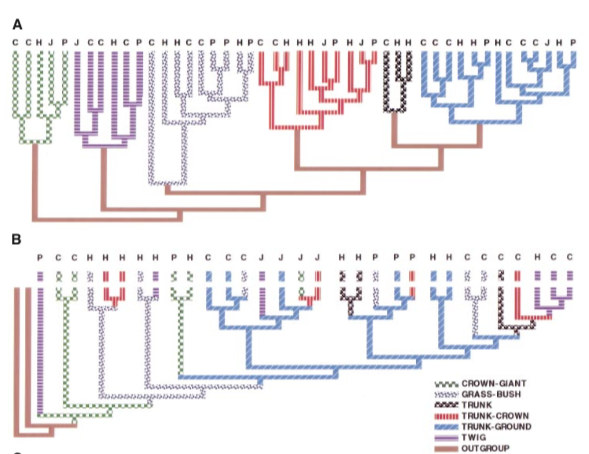
A: Functional dendrogram based on ecomorph
B: Phylogeny indicates frequent evolution of traits
They do not match at all (!!)
Phylogenetically similar = Functional (ecologically) similar??
Putting traits on the tips of phylogeny: phylogenetic signal
[1] Blomberg et al. 2003, Evolution
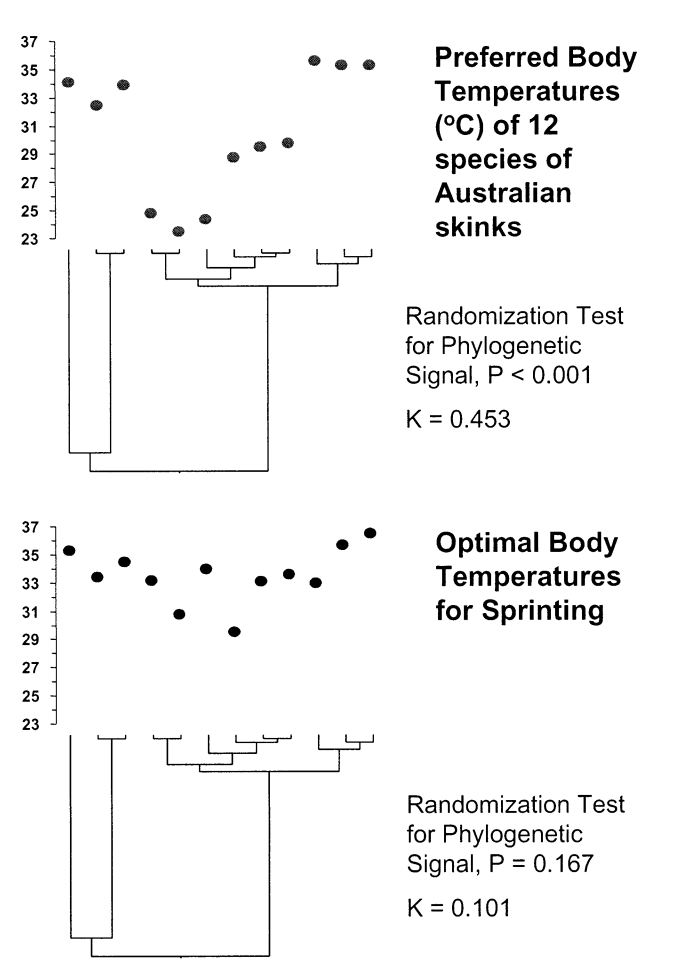
- What is Phylogenetic Signal (K)?
- Phylogenetic signal measures the degree to which related species share similar traits, quantifying the inheritance of traits from either recent or more ancient common ancestors.
- Pagel’s \(\lambda\) is another common measure.
- Interpretation of K Values:
- Large K (phylogenetic conservatism): Trait similarity is high among closely related species, suggesting that traits are conserved across lineages.
- K = 1: Traits are evolving under Brownian motion.
- Small K (phylogenetic divergence): Trait similarity is low among closely related species, indicating that traits have diverged.
- Calculating K:
- K is the ratio of the Mean Squared Error (MSE) of observed trait values to the MSE expected under Brownian motion (random evolutionary change).
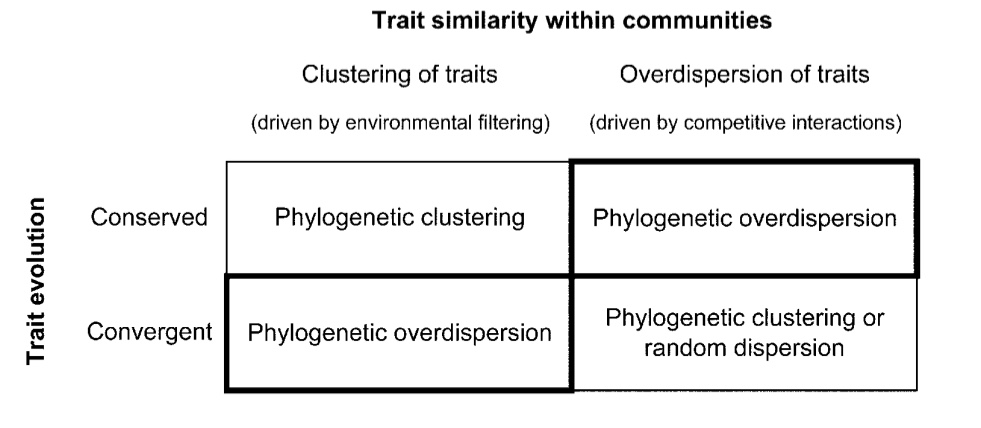
Phylogenetic conservatism matters
[1] Cavender-Bares et al. 2004, The American Naturalist
Phylogenetic conservatism matters
[1] Cavender-Bares et al. 2004, The American Naturalist
The phylogenetic middleman problem
[1] Swenson 2013, Ecography
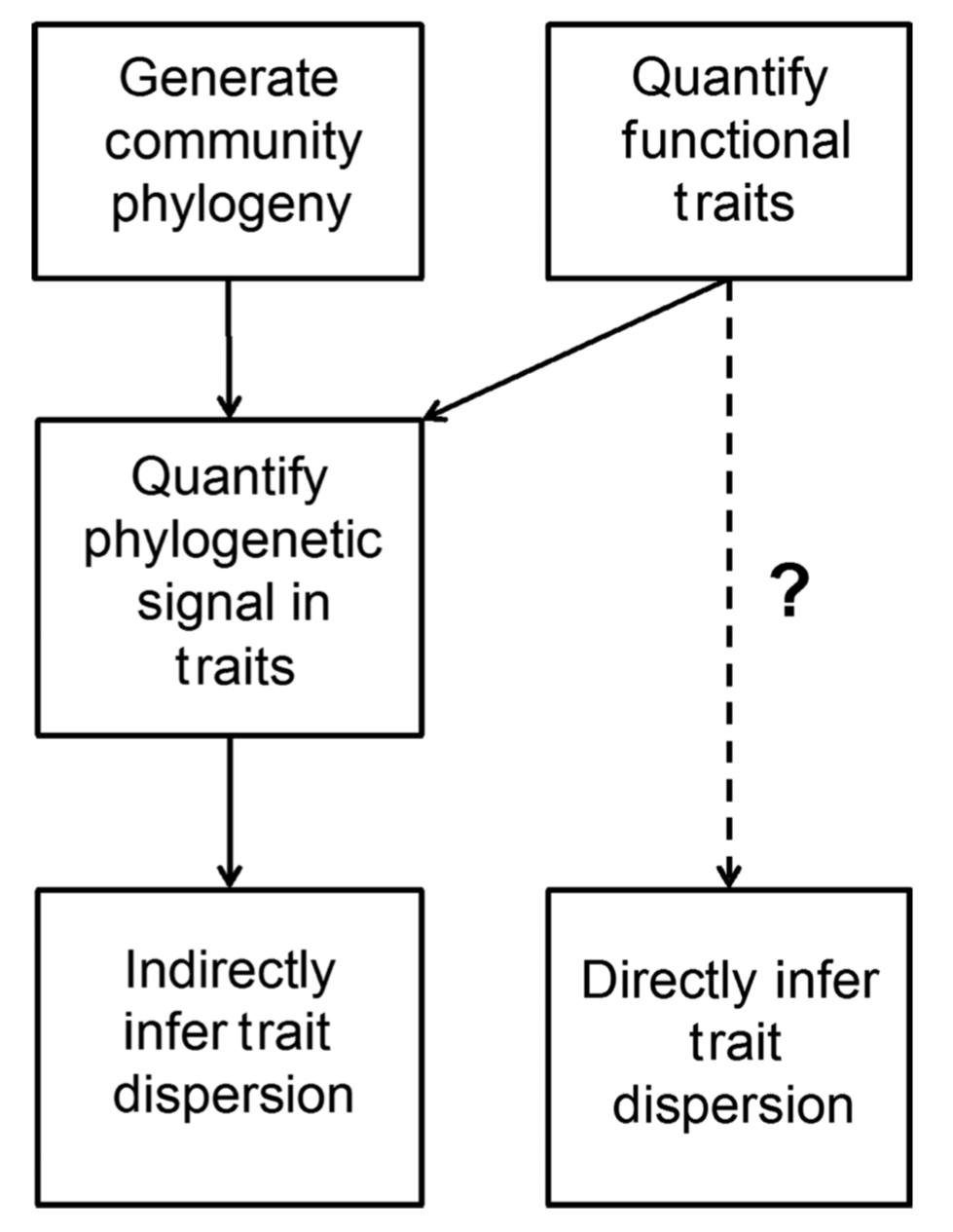
Phylogeny as a proxy for the functional or ecological similarity of species.
Measuring trait data and arraying it on the phylogenetic tree to demonstrate phylogenetic signal in function so that their phylogenetically-based inferences could be supported.
Compared to simply measuring the trait dispersion, this approach is very indirect.
This approach should be avoided! (phylogeny and traits are useful to make meaningful evolutionary inferences)
Plant functional traits
Measurable properties of plants that are indicative of ecological strategies
“Hard” traits: e.g., Photosynthetic rates

“Soft” traits: e.g., LMA (leaf mass per area)

Leaf Economic Spectrum (LES)
[1] Reich et al. 1997, PNAS
[2] Wright et al. 2004, Nature
[3] Osnas et al. 2013, Science
[4] Katabuchi et al. 2025, Oecologia
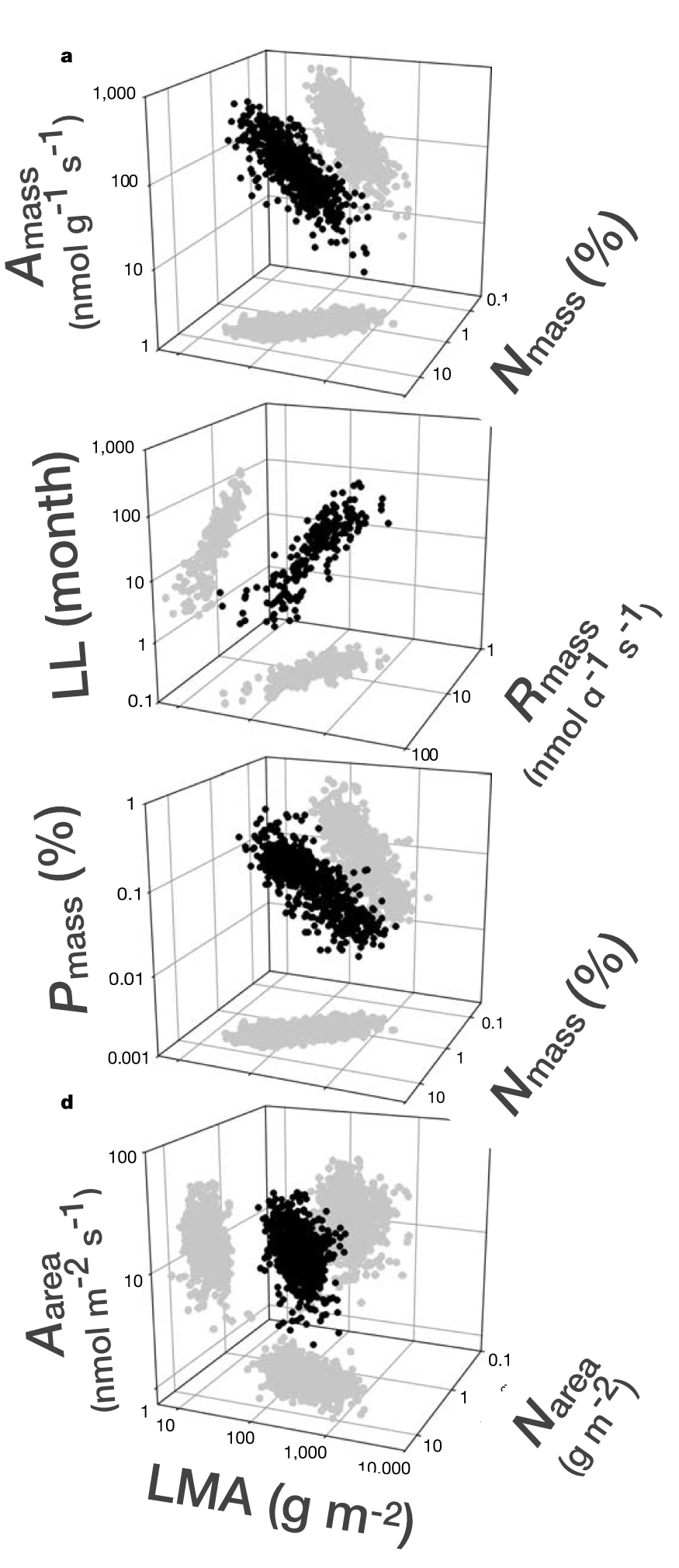
- LES describes pairwise correlations among a bunch of leaf traits from the global leaf database called GLOPNET
- Global leaf function constrained to a single axis (75 % of the variation in the 6 traits)
- Multidimensional (leaf) functional diversity can be mapped into a one-dimensional index
- Controversial because global among‑species patterns don’t always match within‑species or local patterns
Rebuilding community ecology from functional traits
[1] McGill et al. 2006, Trends in Ecology and Evolution
Non-trait based statement
- Campanula aparinoides is found only in infertile habitats.
Trait-based statement
- Compact plants with canopy area < 30 cm 2 and small or absent leaves are restricted to marshes with < 18 \(\mu\) g g -1 soil P.
Rebuilding community ecology from functional traits
[1] McGill et al. 2006, Trends in Ecology and Evolution
- Go beyond ‘How many species and why?’ to ask ‘How much variation in traits and why?’
- Go beyond ‘In what environments does a species occur?’ to ask ‘What traits and environmental variables are most important in determining fundamental niche?’
- Go beyond ‘What are the most important niche dimensions?’ to ask ‘What traits are most decisive in translating from fundamental niche to realized niche?’
- Go beyond ‘How does population dynamics determine abundance?’ to ask ‘How does the performance of species in the interaction milieu determine their ranking of abundance or biomass?’
- Go beyond ‘How does space affect population dynamics?’ to ask ‘How do environmental gradients affect community structuring?’
Convex hull volume (functional richness)
[1] Cornwell et al. 2006, Ecology
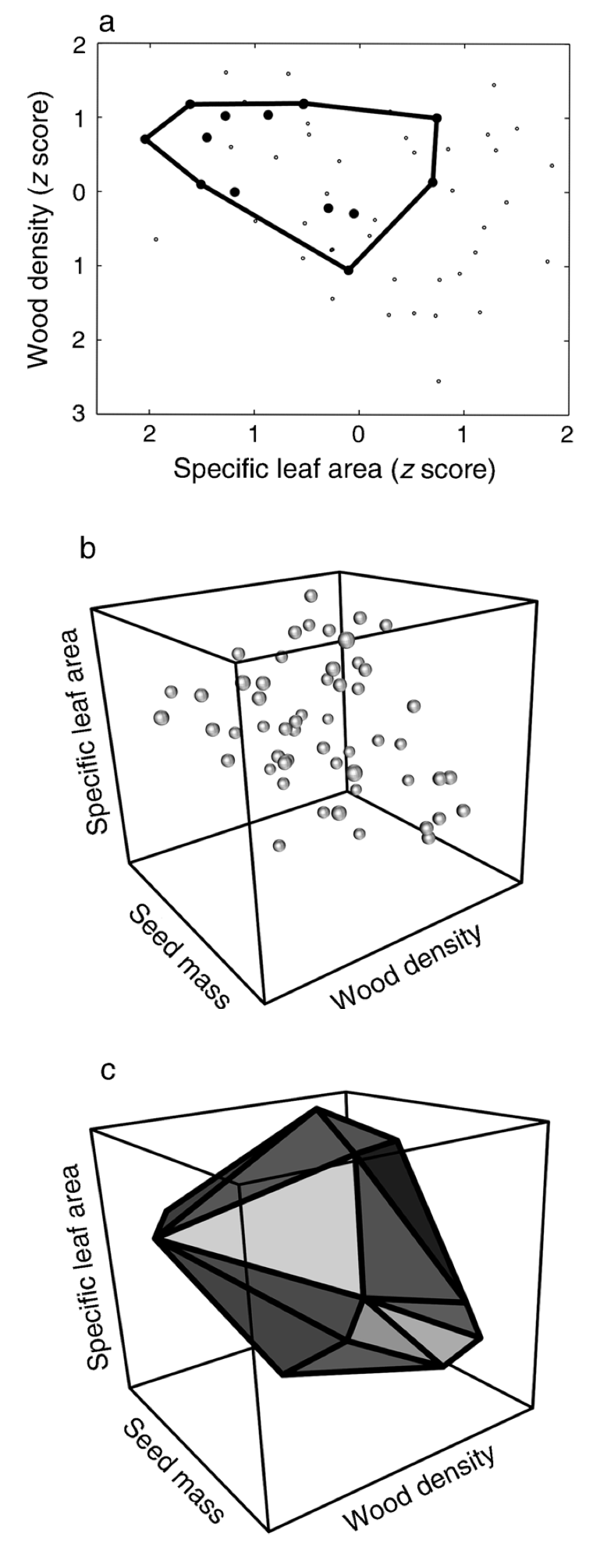
California woody-plant communities (43 plots, 54 species, 3 traits)
Is the trait volume of California woody-plant communities significantly less than expected by chance?
- Environmental filtering
Convex hull volume (functional richness)
[1] Cornwell et al. 2006, Ecology
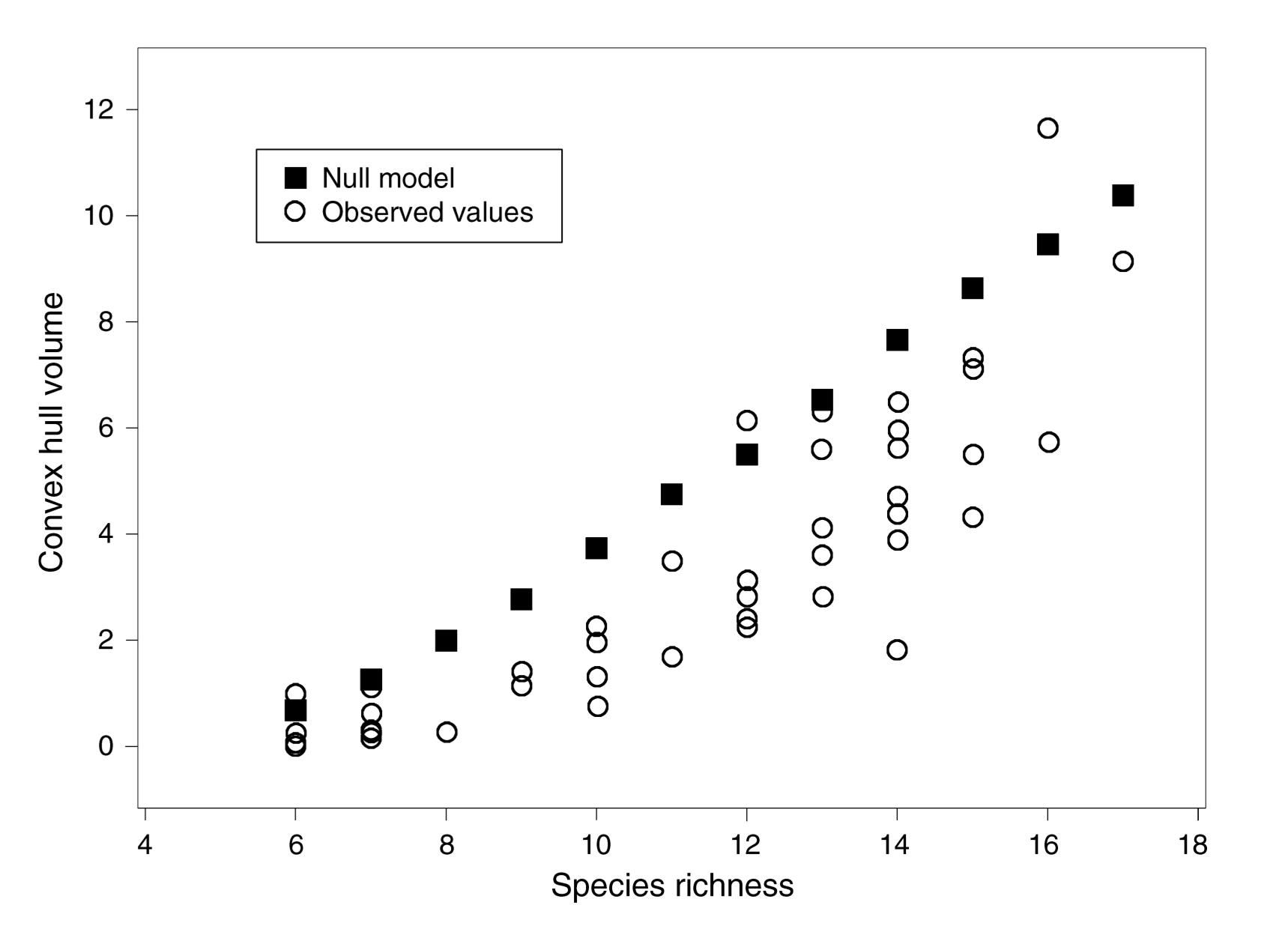
Species in 40 out of 43 plots occupied less trait space than would be expected by chance
Consistent with environmental filtering
Community assembly and trait distribution
[1] Cornwell & Ackerly 2009, Ecological Monographs
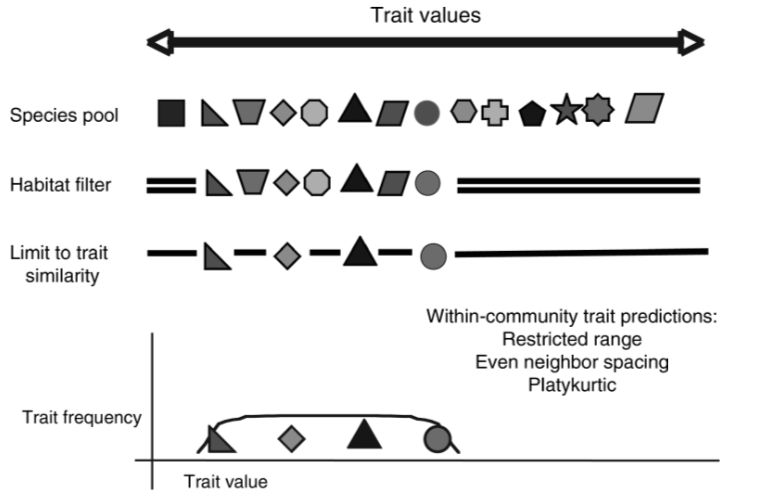
Environmental filtering and limiting similarity can occur at the same time
[1] Kraft et al. 2008, Science
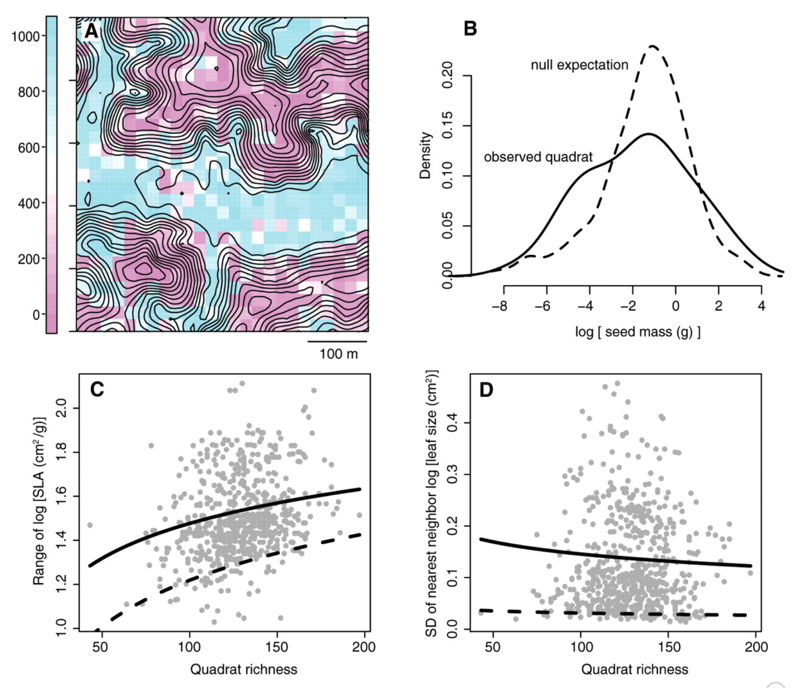
Yasuni tropical tree communities, 25ha, 625 20m x 20m quadrats, 1089 species!
Consistent with environmental filtering
A: Ridgetops have lower than expected SLA and valleys have higher
- Traits match with environmental conditions
B: Seed mass shows broader distribution than expected - Limiting similarity
C: Range of SLA is smaller than expected - Environmental filtering
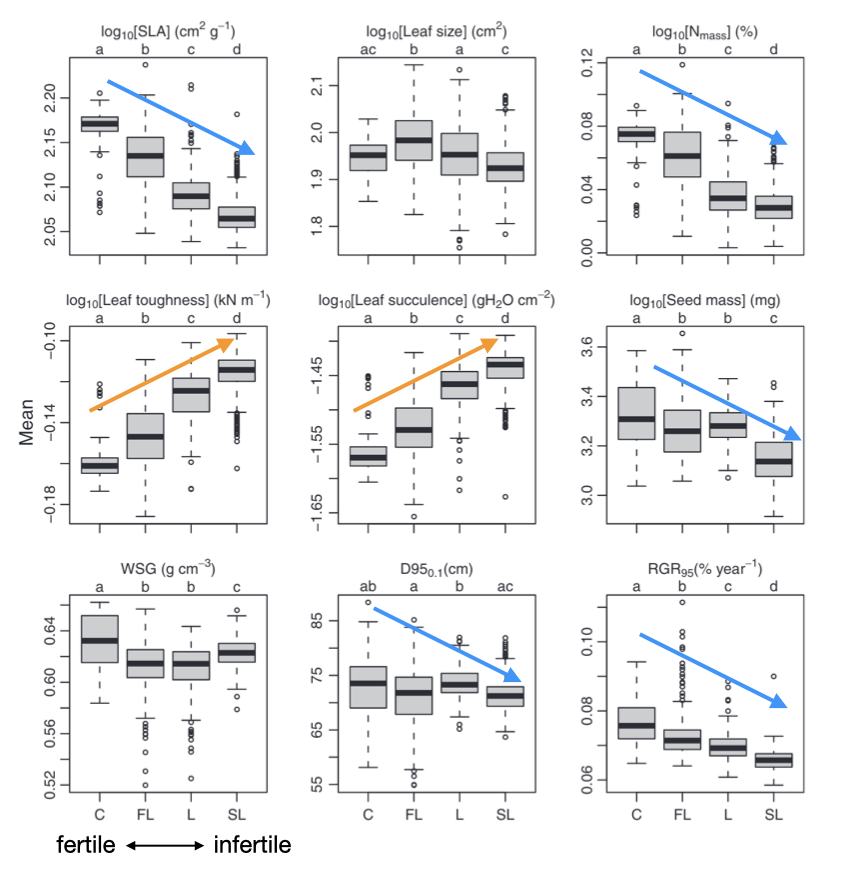
Environmental filtering can occur within dipterocarp trees
[1] Katabuchi et al. 2012, Journal of Ecology
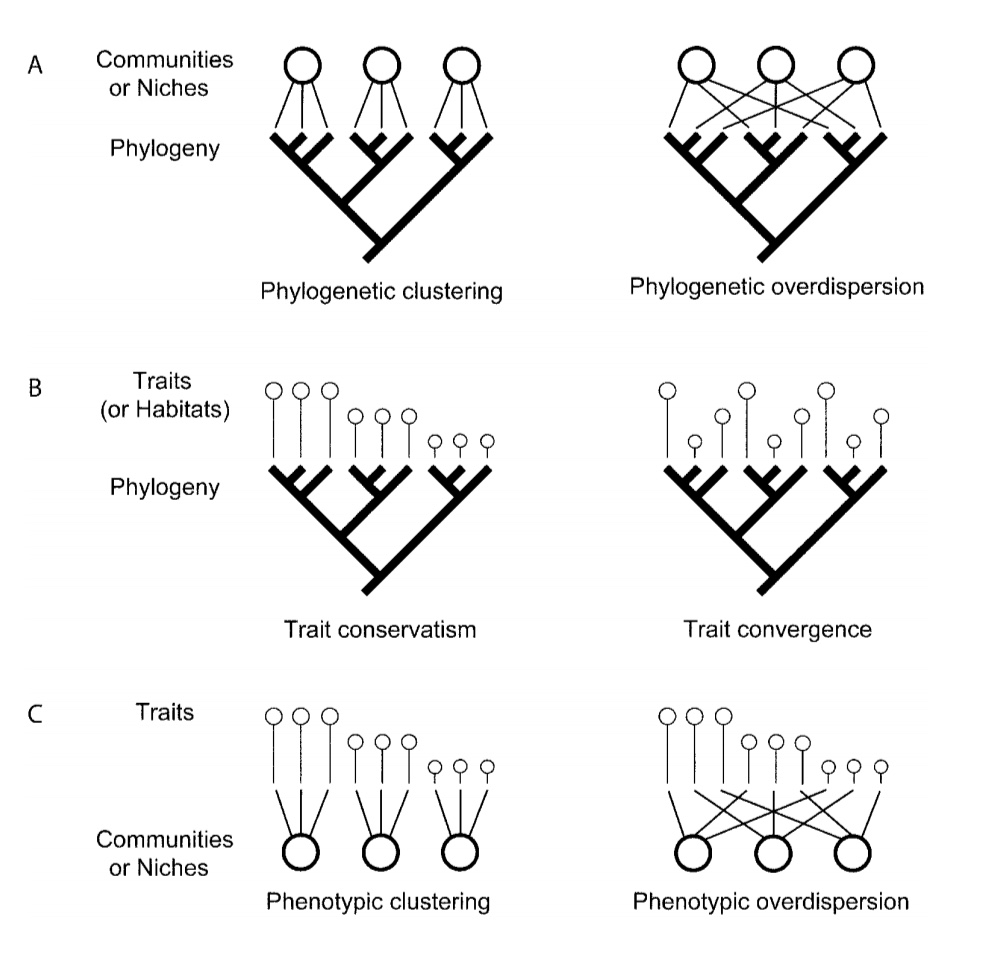
How ecological processes might influence community assembly
| Process / Factor | Pattern |
|---|---|
| Biotic factor | |
| Environmental filtering | Clustering |
| Abiotic factor (competition) | |
| Limiting similarity / Competitive exclusion | Overdispersion |
| Competitive hierarchy / directional competition | Clustering |
| Herbivores / Parasites / Pathogens | Overdispersion |
| Pollinator-mediated competition | Overdispersion |
| Abiotic factor (facilitation) | |
| Nurse plants | Overdispersion |
| Pollinator facilitation | Clustering |
| Stochastic process | |
| Neutral theory | Random |
Competitive hierarchy
[1] \(t_A\) and \(t_B\) are the functional trait values of species A and B
[2] Kunstler et al. 2012, Ecology Letters

Limiting similarity
Competitive interaction strengths between species will increase with decreasing niche distance, measured as their absolute traits distance \(|t_A - t_B|\)

Competitive hierarchy
Competitive effects of species A on species B will increase with increasing \(t_A - t_B\).
Plant–herbivore interactions
[1] Endara et al. 2022, Journal of Ecology
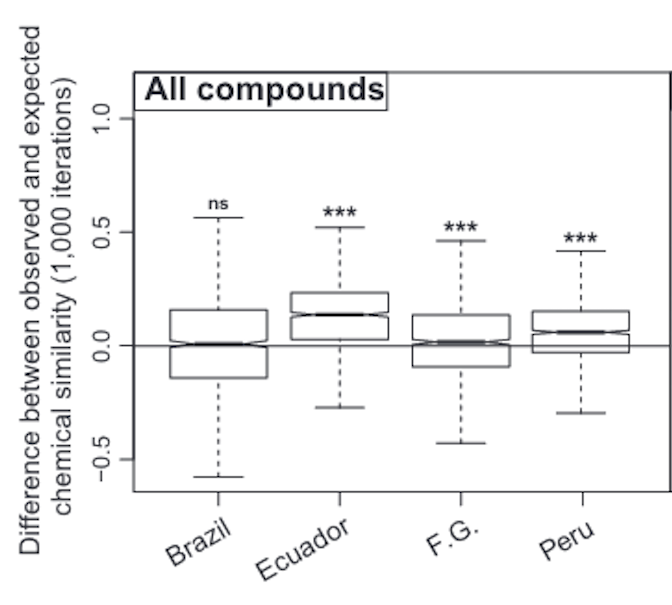
6000+ secondary metabolites from nearly 100 species in a diverse Neotropical plant clade across the whole Amazonia
More differences in their defensive chemistry than expected by chance
Plant–herbivore interactions promote species diversity
Facilitation
[1] López-Angulo et al. 2018, Journal of Vegetation Science
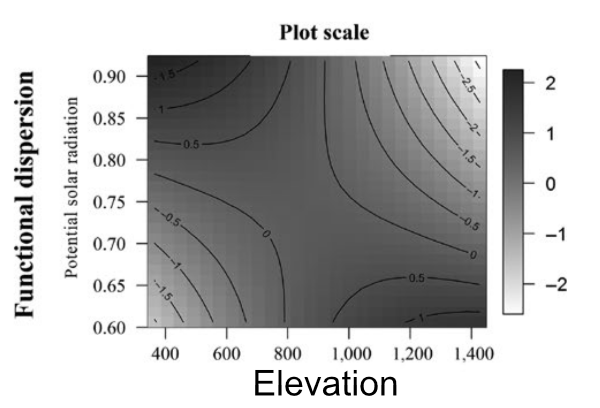
Alpine plants in the Andes
Functional dispersion in harsh environments (higher potential solar radiation)
Facilitation tends to dominate interactions when environmental harshness increases
Trait-based ecology: where are we now?

Community averages can mislead without species‑level context
[1] Katabuchi et al. 2017, Ecology
[2] Swenson et al. 2020, Ecological Monographs
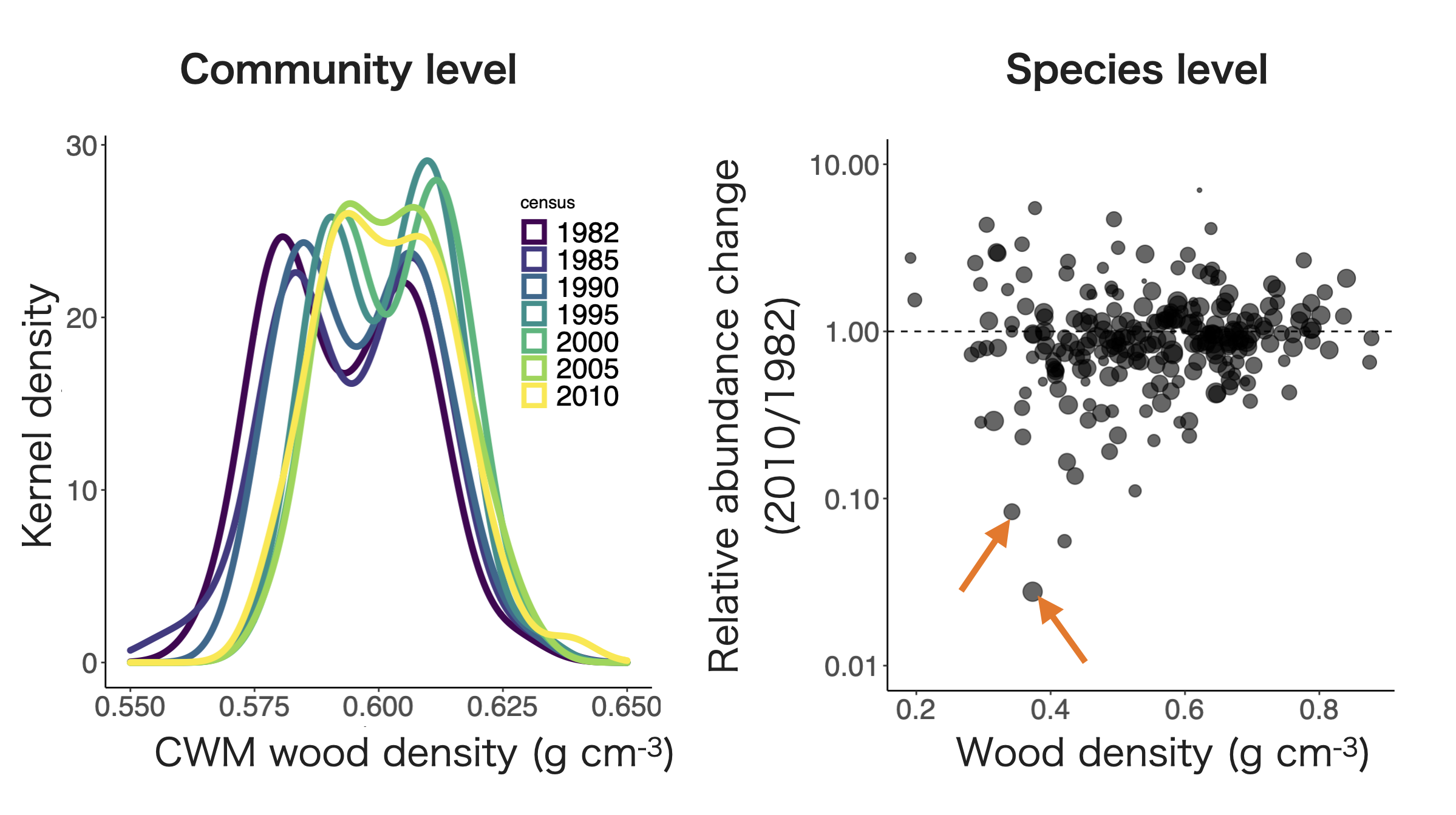
50ha Forest Dynamics Plot on Barro Colorado Island, Panama
CWM wood density trends suggest a community-level response in climate change (i.e, drier conditions).
But 2 of ~300 species (~0.7%) account for ~60% of the temporal shifts in CWM, likely due to species-specific pathogens rather than a community-wide climate response.
Inter‑ and intra‑specific trait patterns often diverge
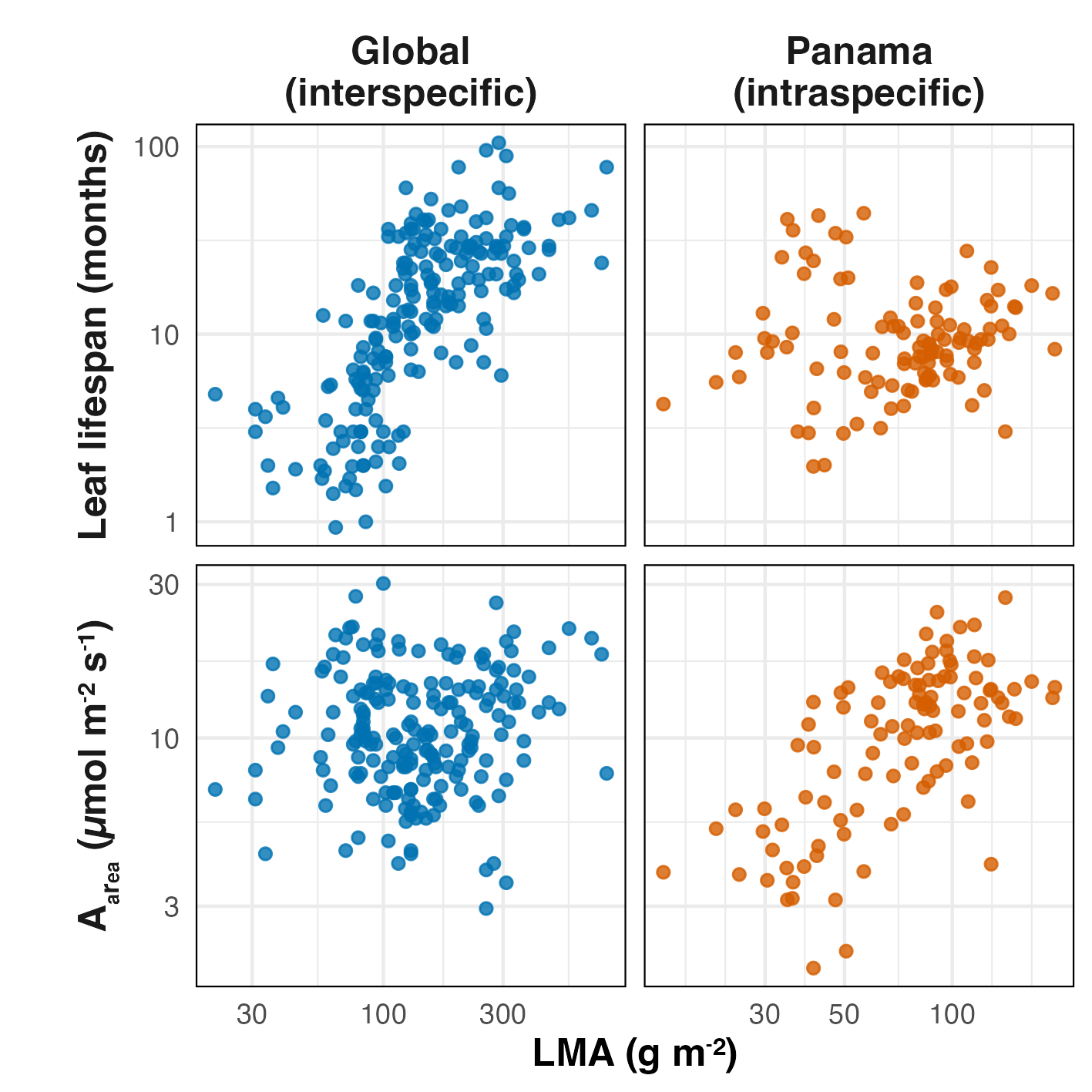
[1] Osnas, Katabuchi et al. 2018, PNAS
Using harder-to-measure physiological traits may not be enough
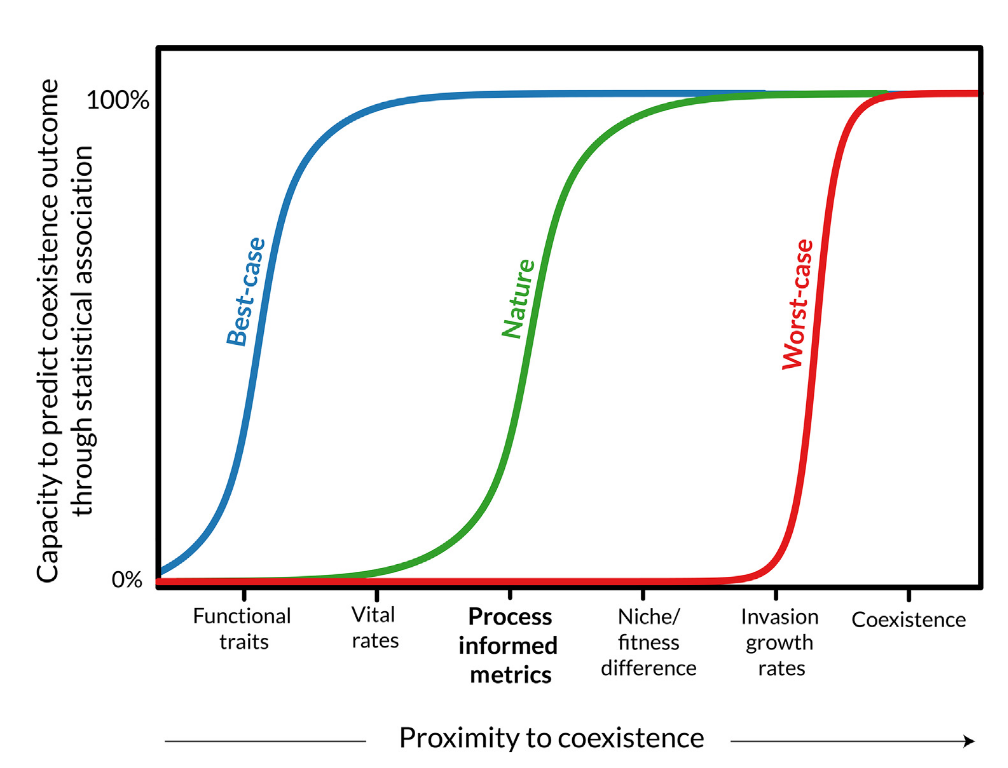
We may need metrics somewhere between functional traits and invasion growth rate (model parameter) to predict coexistence.
[1] Levine et al. 2024, Trends in Ecology & Evolution
Decomposing LMA into metabolic (LMAm) and structural (LMAs) components improves physiological prediction
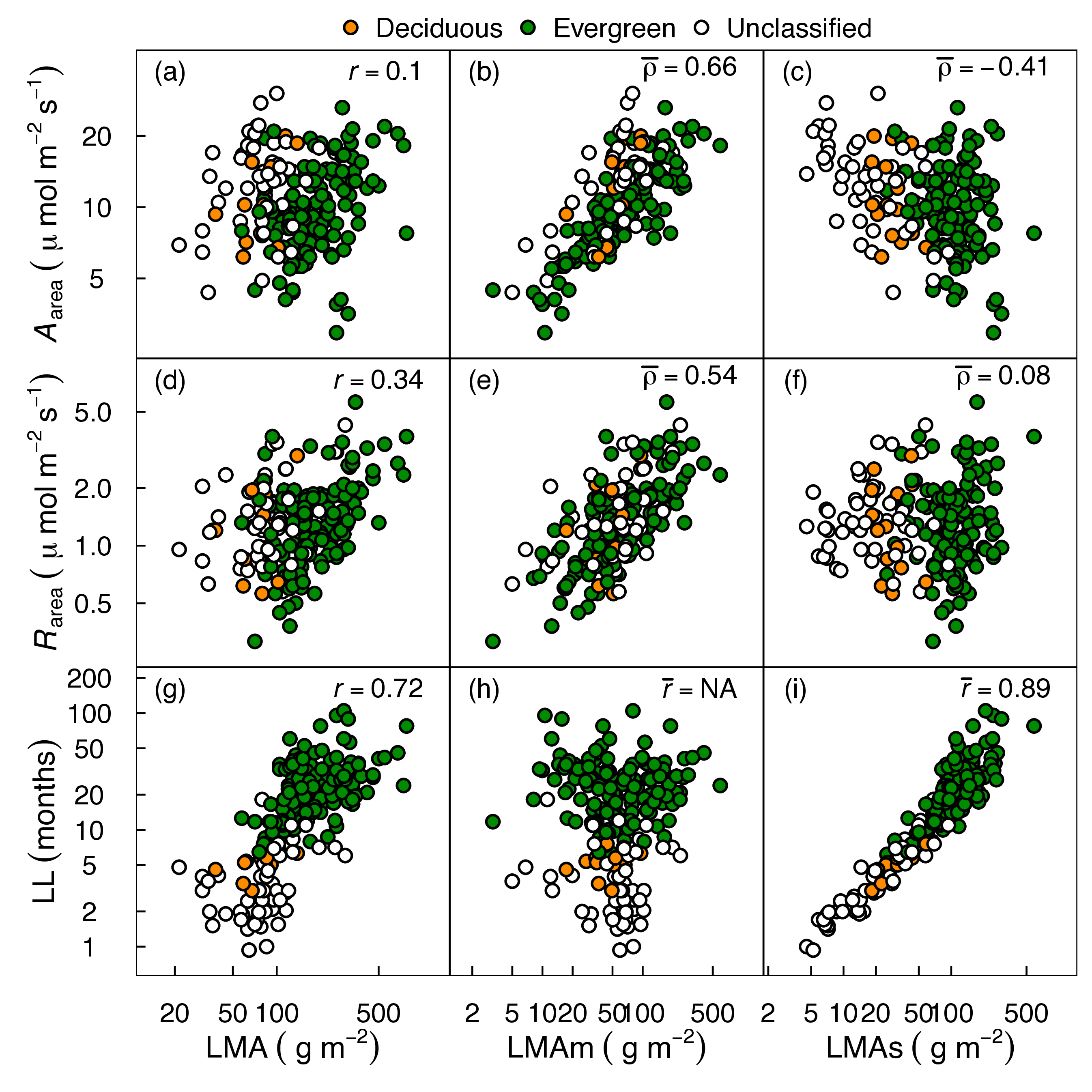
PIMs (LMAm/LMAs) outperform bulk LMA for predicting physiology
Katabuchi et al. 2025, Oecologia
Summary 🍺
Why do we use trait and phylogenetic diversity?
We want to quantify ecological similarities and biodiversity dimensions.
We need to be very careful when we use or develop ways to map multiple dimensions of biodiversity to a few dimensions of diversity.
Trait-based approaches
Moving ecology towards more quantitative and predictive methods.
We only focused on a “snapshot” of biodiversity (predicting future biodiversity is another story)
Developing more effective metrics may be necessary, beyond simply measuring traits.
How to calculate trait and phylogenetic diversity anyway?
- We have some R practice.
About the slides
The slides are made using Quarto with Emi Tanaka’s CSS design and Danyang Dai’s template.
Source code for the slides can be found at mattocci27/phy-fun-div
